Applications are closed but you can express your interest in the next edition by clicking on the button above and filling in the form.
This is a Cajal NeuroKit course that combines online lectures about fundamentals and advanced neuroscience topics with hands-on and physical experiments.
Researchers can participate from anywhere in the world because the course material is shipped to participants in a kit box that contains all the tools needed to follow the online course.
This course is now at its third edition. It is held at least once a year.
Call for TAs: If you already know how to programme in Bonsai and you would like to run this course locally as a Teaching Assistant with a group of students, please click on the button below to fill in the TA form. Thank you.
Course overview
Modern neuroscience relies on the combination of multiple technologies to record precise measurements of neural activity and behaviour. Commercially available software for sampling and controlling data acquisition is often too expensive, closed to modification and incompatible with this growing complexity, requiring experimenters to constantly patch together diverse pieces of software.
This course will introduce the basics of the Bonsai programming language, a high-performance, easy to use, and flexible visual environment for designing closed-loop neuroscience experiments combining physiology and behaviour data.
This language has allowed scientists with no previous programming experience to quickly develop and scale-up experimental rigs, and can be used to integrate new open-source hardware and software.

Course Teaser
What will you learn?
By the end of the course you will be able to use Bonsai to:
– create data acquisition and processing pipelines for video and visual stimulation.
– control behavioral task states and run your closed-loop experiments.
– collect data from cameras, microphones, Arduino boards, electrophysiology devices, etc.
– achieve precise synchronization of independent data streams.
The online material will be soon found here.
Faculty

Gonçalo Lopes
Course Director
NeuroGEARS, London, UK
Instructors
João Frazão – Champalimaud Research, Lisbon, PT
Niccolò Bonacchi – International Brain Laboratory, Lisbon, PT
Nicholas Guilbeault – University of Toronto, CA
André Almeida – NeuroGEARS, London, UK
Bruno Cruz – NeuroGEARS, London, UK
Programme
Day 1 – Introduction to Bonsai
-
Introduction to Bonsai. What is visual reactive programming.
-
How to measure almost anything with Bonsai (from quantities to bytes).
-
How to control almost anything with Bonsai (from bytes to effects).
-
How to measure/control multiple things at the same time with one computer.
-
Demos and applications: a whirlwind tour of Bonsai.
Day 2 – Cameras, tracking, controllers
-
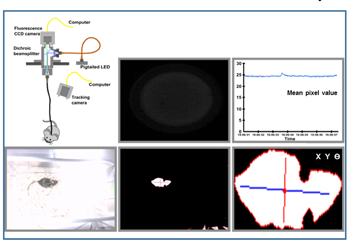
Measuring behavior using a video.
-
Recording real-time video from multiple cameras.
-
Real-time tracking of colored objects, moving objects and contrasting objects.
-
Measuring behavior using voltages and Arduino.
-
Data synchronization. What frame did the light turn on?
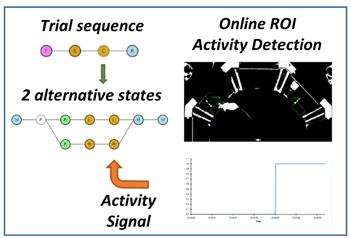
Day 3 – Real-time closed-loop assays
-
What can we learn from closed-loop experiments?
-
Conditional effects. Triggering a stimulus based on video activity.
-
Continuous feedback. Modulate stimulus intensity with speed or distance.
-
Feedback stabilization. Record video centered around a moving object.
-
Measuring closed-loop latency.
Day 4 – Operant behavior tasks
-
Modeling trial sequences: states, events, and side-effects.
-
Driving state transitions with external inputs.
-
Choice, timeouts and conditional logic: the basic building blocks of reaction time, Go/No-Go and 2AFC tasks.
-
Combining real-time and non real-time logic for good measure.
-
Student project brainstorming
Day 5 – Visual stimulation and beyond
-
Interactive visual environments using BonVision.
-
Machine learning for markerless pose estimation using DeepLabCut.
-
Multi-animal tracking and body part feature extraction with BonZeb.
-
Student project presentation.
-
Where to next.
The course will be held from 13:00 to 17:00 GMT.
Registration
Registration fee: 500€ per person (includes shipping of the course kit, pre-recorded and live lectures before and during the course, full attendance to the course, and course certificate).
The CAJAL programme can offer some stipends (waived partial or full registration fee). Please apply through the course online application form.
Registration fee for a group: 500€ for one person and one course kit + 150€ for any additional person (without the course kit)
Applications are closed but you can express your interest in the next edition by clicking on the button above and filling in the form.
Please note that this is not considered as an application.
To receive more information about this NeuroKit, email info@cajal-training.org









